Introducing Tom Miller
Tom Miller is a newly appointed Associate Professor at the Center for Chromosome Stability in the Department of Cellular and Molecular Medicine. ISBUC recently sat down with Tom and found out all about his approach to cryo-EM and his research into the mechanisms of eukaryotic chromosome replication.

What are the big research questions you are working on?
Our lab is interested in the mechanisms of eukaryotic chromosome replication. Often we talk about DNA-replication but chromosomes are so much more complex than just DNA. Eukaryotic genomes are covered in all sorts of proteins. Some, like histones, are neatly redistributed after replication but others can interfere with replication and are a threat to genome stability. DNA damage can also block the replication machinery. So, the question we are interested in is how the replication machinery replicates chromosomes rather than just DNA, and how it does this accurately.
How did you become interested in these questions?
Since my undergraduate studies, I have always had an interest in both structural biology and chromatin biology. After a postdoc at the EMBL in Heidelberg, I had the opportunity to join Alessandro Costa’s lab, just as he was moving to The Francis Crick Institute. Alessandro’s group were at the forefront of studying eukaryotic DNA replication using cryo-EM, at a time when the cryo-EM ‘resolution revolution’ was really taking off. I joined Alessandro’s lab to learn cryo-EM and use it to study how chromatin is replicated.
With the goal of reconstituting replication, I initially imaged an MCM helicase loading reaction to see if I’d been able to perform the first step in assembling the replication machinery. At the microscope Alessandro and I immediately realized that we were looking at something exciting and that we were going to be able to answer all of these questions about helicase loading that we hadn’t originally intended on looking into. Literally, we looked down the microscope and thought what is that! And so the mechanism of MCM loading became my postdoc project.
What was it like working at The Francis Crick Institute?
The Crick is a great place to work. It is filled with amazing researchers and great facilities. In particular, the core Science Technology Platforms and the people that run them are fantastic. One of the things that I really liked was the large open-plan lab spaces, which were shared by about six different research groups. Each group would have its own dedicated bench space but in the centre there would be all the communal equipment. This meant that you ended up sharing your labs with other groups who would be working on quite different areas of biology. This created a very collaborative and open environment.
What is the visual biochemistry technique that you have pioneered?
The visual biochemistry approach is a simple idea that takes advantage of the unique capabilities of cryo-EM. Rather than imaging a purified homogeneous sample trapped in a particular state, we look at complete biochemical reactions, as they occur over time. This allows us to find all kinds of novel intermediate states that might otherwise be very difficult to identify.
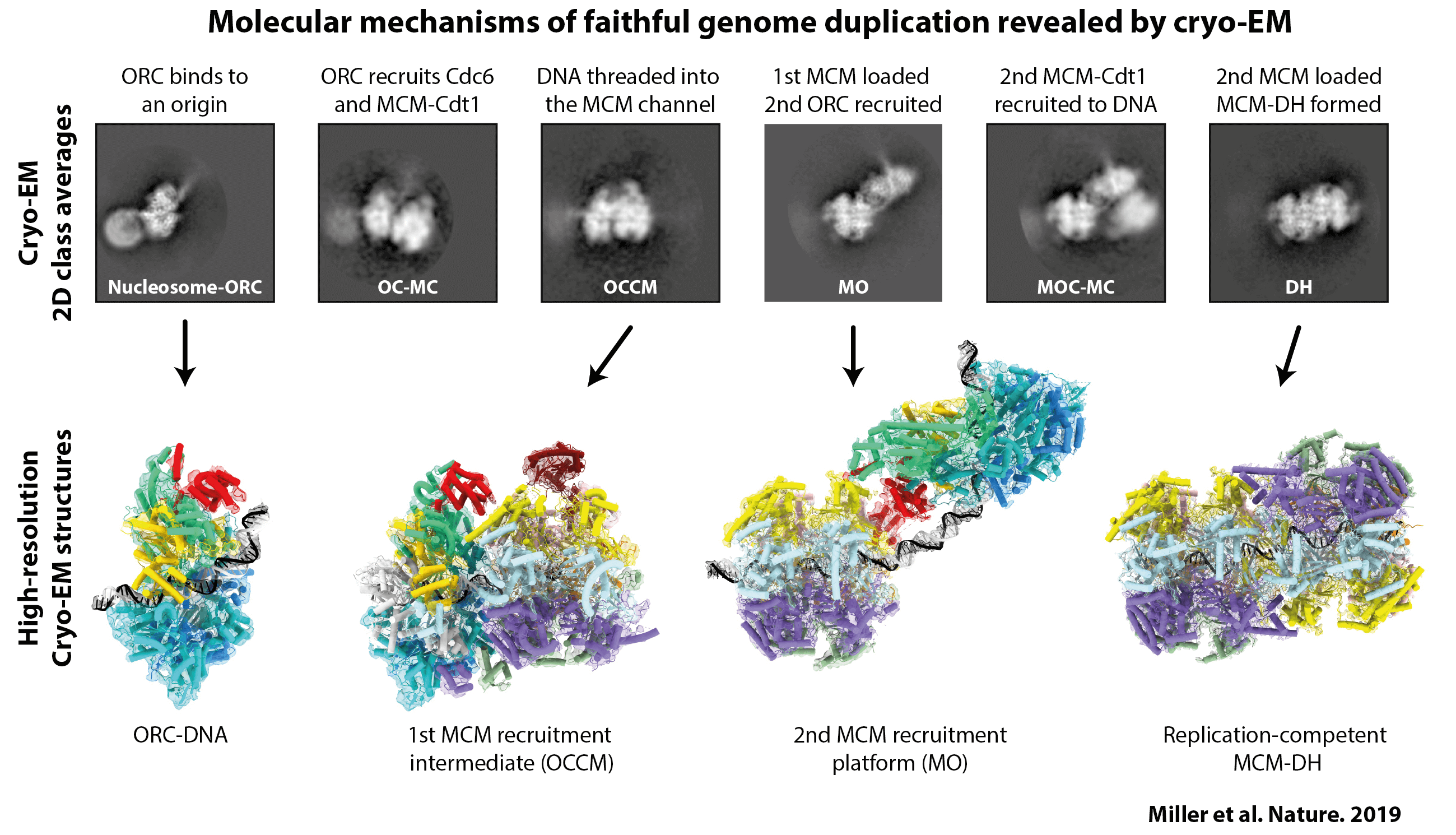 I first applied this approach to the process of MCM helicase loading. We had already captured some high-resolution images of specific states in the helicase loading process—either by looking at individual factors, the purified end-point of the reaction or by using nucleotide analogues that stall the reaction—but there were still several critical steps missing. To try to find them, we mixed all of the components required for loading the MCM helicase: the proteins were wild type, we didn’t use cross-linkers and the reactions were performed in the presence of ATP. We then imaged the reaction at different time points and monitored the change in populations of the different intermediates. Through this method, we found multiple novel intermediate states (as demonstrated in the above figure). One in particular gave us a lot of insight into the mechanism of MCM helicase loading and reconciled a lot of apparently conflicting data in the field. The idea now is to apply this approach to later steps in the chromosome replication pathway.
I first applied this approach to the process of MCM helicase loading. We had already captured some high-resolution images of specific states in the helicase loading process—either by looking at individual factors, the purified end-point of the reaction or by using nucleotide analogues that stall the reaction—but there were still several critical steps missing. To try to find them, we mixed all of the components required for loading the MCM helicase: the proteins were wild type, we didn’t use cross-linkers and the reactions were performed in the presence of ATP. We then imaged the reaction at different time points and monitored the change in populations of the different intermediates. Through this method, we found multiple novel intermediate states (as demonstrated in the above figure). One in particular gave us a lot of insight into the mechanism of MCM helicase loading and reconciled a lot of apparently conflicting data in the field. The idea now is to apply this approach to later steps in the chromosome replication pathway.
One other tool that we developed as a part of the visual biochemistry approach involves what we call ‘reconstitution in silico’. Up until now, when you have imaged multiple proteins or complexes that are associated with a flexible substrate, you usually have no idea where on the substrate the protein complexes sit or how they relate to one another. We developed an approach for finding this out. For example, when we were looking at helicase loading, this approach allowed us to see where the first and then second MCM helicase are initially loaded onto the DNA, and allowed us to see that MCM sliding was a key part of the mechanism. This approach has obvious value when studying complexes attached to DNA but it could also be used to study the relationships between any factors that are flexibly associated – for example two proteins in a membrane.
How has the transition to Copenhagen been?
Copenhagen is a great city and the chromosome biology community here is very strong. I have been really impressed by the way the community comes together from all of the different University departments and research centres. People actually get together, talk and collaborate, it is really nice.
I am based at the Center for Chromosome Stability and everybody there has been very welcoming and helpful. It is a great tight-knit community of researchers working on similar topics. There is also the Chromosome Biology Club. I gave a talk there in my first few weeks in Copenhagen and immediately, this sparked new connections and opportunities for collaboration. For example, this led to an opportunity to work with Hana Sedlackova in Jiri Lukas’ group at the CPR, which has been really enjoyable.

And how is it going setting up your lab?
It is going well. Fortunately, I have had lots of support the members of the CCS and from members of the CPR including Julian Duxin, Niels Mailand, Guillermo Montoya, Eva Kummer, Nicholas Taylor and Tillman Pape who have helped us get going. I have recruited postdoc Christl Gaubitz, who comes from Brian Kelch’s lab at the University of Massachusetts Medical School, where she has been working on DNA replication. I have also recruited a PhD student, Sarah Frederiksen Ruidiaz, who has come from Birthe Kragelund’s lab here in Copenhagen. We’ve also been working with two lab technicians, Carina Lynnerup Pedersen and Alex Avram, who joined us from within the ICMM. They are getting the biochemistry set up in the lab, purifying proteins and starting to do some experiments. It is very exciting and I look forward to seeing what we can achieve as a team!
Where can we learn more about your research?
My research on MCM helicase loading was published in: Miller, T.C.R., Locke, J., Greiwe, J.F. et al. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 575, 704–710 (2019).
For more information on the ‘reconstitution in silico’ approach see our recent Current Opinion article: Greiwe, J. F., Zanetti, G., Miller, T. C. R. & Costa, A. In silico reconstitution of DNA replication. Lessons from single-molecule imaging and cryo-tomography applied to single-particle cryo-EM. Curr Opin Struct Biol 72, 279-286 (2022).
